Abstract
Fighting fish species in the genus Betta are found in several Southeast Asian countries. Depending on the mode of paternal care for fertilized eggs and hatchlings, various species of the betta fish are classified as mouth brooders or nest builders whose members in turn have been grouped according to their similarities mainly in morphology.
The mouth brooders as well as some nest builders involved in the present study include fishes discovered and identified subsequent to previous reports on species groupings and their positions on phylogenetic trees based on DNA sequences that differ from those used by us in this study. From the mitochondrial COI gene and nuclear ITS1 gene sequences and more accurate analyses we conclude that the following members of the mouth-brooding pairs, named differently previously, are virtually identical, viz the Betta prima–Betta pallida pair and Betta ferox–Betta apollon pair. The Betta simplex, hitherto believed to be one species, could possibly be genetically split into 2 distinct species.
In addition, several other established type-locality fishes could harbor cryptic species as judged by genetic differences. Assignments of fish species to groups reported earlier may have to be altered somewhat by the present genetic findings. We propose here a new Betta fish phylogenetic tree which, albeit being similar to the previous ones, is clearly different from them. Our gene-based evidence also leads to assignments of some fishes to new species groups and alters the positions of some species on the new phylogenetic tree, thus implying different ancestral relationships.
Keywords: Betta fish species, Bubble-nest builder, Cryptic species, Fighting fish, Mouth brooder, Phylogenetic tree, Sister group, Southeast Asia
Introduction
Fighting fishes in the genus Betta have received constant international interest in terms of discovery and identification of species. These fishes have been reported in Cambodia, Indonesia, Lao PDR, Malaysia, Myanmar, Singapore, Thailand and Vietnam (Kowasupat et al., 2012a, Kowasupat et al., 2012b, Schindler and Linke, 2013, Schindler and Schmidt, 2006, Schindler and Schmidt, 2008, Tan, 2009a, Tan, 2009b, Tan, 2013, Tan and Ng, 2005, Witte and Schmidt, 1992). In caring for fertilized eggs and newly hatched fry, the male of one type of fish (the bubble nester) builds a bubble nest as temporary shelter for the vulnerable ones, whereas the male of the other type (the mouth brooder) protects the fertilized eggs and the hatchlings in its mouth until release. Generally male bubble nesters are more colorful than male mouth brooders with a few exceptions. Some of the better known bubble nesters are highly colorful and easy to identify, viz Betta splendens, Betta imbellis, Betta smaragdina, Betta mahachaiensis, Betta coccina and Betta livida. Among the mouth brooders, some of the species are, e.g., Betta apollon, Betta chloropharynx, Betta pi, Betta prima, Betta pugnax, Betta simplex and Betta stigmosa. Table 1 lists the two types of betta fishes mentioned in this article according to paternal care and species groups proposed earlier (Kowasupat et al., 2014, Schindler and Schmidt, 2006, Tan and Ng, 2005, Witte and Schmidt, 1992). Identifications of these species have been based solely on morphological criteria in a large number of cases.
Table 1.
Groupings of fighting fish species used in this study. (The betta species grouped here are as in Witte and Schmidt (1992), Tan and Ng (2005), Schindler and Schmidt (2006), and Kowasupat et al. (2014).).
| Species group | Species | Country | Caught (C)/Purchased (P) |
|---|---|---|---|
| Mouth-brooding betas | |||
| B. dimidiata group | B. krataios Tan and Ng, 2006⁎ | I | P |
| B. foerschi group | B. rubra Perugia, 1893⁎ | I | P |
| B. picta group | B, edithae Vierke, 1984 | I | P |
| B. simplexKottelat (1994) | T | C | |
| B. sp. (cf. simplex)⁎ | T | C | |
| B. primaKottelat (1994)⁎ | T & Cm | C | |
| B. sp. (cf. prima)⁎ | V | P | |
| B. enisae Kottelat, 1995⁎ | I | P | |
| B. pallida Schindler and Schmidt, 2004⁎ | T | C | |
| B. pugnax group | B. pugnax Cantor, 1850 | M | C |
| B. sp. (cf. pugnax)⁎ | M | C | |
| B. pulchra Tan and Tan, 1996⁎ | M | C | |
| B. sp. (cf. pulchra)⁎ | M | C | |
| B. stigmosaTan and Ng (2005)⁎ | M | C | |
| B. apollonSchindler and Schmidt (2006)⁎ | T | C | |
| B. sp. (cf. apollon) 1⁎ | T | C | |
| B. sp. (cf. apollon) 2⁎ | T | C | |
| B. feroxSchindler and Schmidt (2006)⁎ | T | C | |
| B. sp. (cf. ferox)⁎ | T | C | |
| B. kuehneiSchindler and Schmidt (2008)⁎ | M | C | |
| B. sp. (cf. kuehnei)⁎ | M | C | |
| B. unimaculata group | B. unimaculata Popta, 1905 | I | P |
| B. macrostoma Regan, 1910 | B | P | |
| B. patoti Weber and de Beaufort, 1922 | I | P | |
| B. ocellata de Beaufort, 1933 | M | P | |
| B. ideii Tan and Ng, 2006⁎ | I | P | |
| B. compuncta Tan and Ng, 2006⁎ | I | P | |
| B. waseri group | B. waseri Krummenacher, 1986 | M | C |
| B. hipposideros Ng and Kottelat, 1994 | M | C | |
| B. chloropharynx Kottelat and Ng, 1994 | I | P | |
| B. pi Tan, 1998 | T | P | |
| Bubble-nest building betas | |||
| B. bellica group | B. bellica Sauvage, 1884⁎ | M & I | P |
| B. coccina group | B. coccina Vierke, 1979 | M | C & P |
| B. tussyae Schaller, 1985 | M | C | |
| B. persephone Schaller, 1986⁎ | M | P | |
| B. livida Ng and Kottelat, 1992⁎ | M | C & P | |
| B. smaragdina group | B. smaragdina Ladiges, 1975 | T | C |
| B. sp. (cf. smaragdina) 1 Kowasupat et al. (2014)⁎ | T | C | |
| B. sp. (cf. smaragdina) 3 Kowasupat et al. (2014)⁎ | L | C | |
| B. sp. (cf. smaragdina) 4 Kowasupat et al. (2014)⁎ | T | C | |
| B. stiktosTan and Ng (2005)⁎ | Cm | P | |
| B. splendens group | B. splendens Regan, 1910 | T, L, Cm & My | C & P |
| B. imbellis Ladiges, 1972 | T, M & V | C & P | |
| B. mahachaiensisKowasupat et al., 2012a, Kowasupat et al., 2012b⁎ | T | C & P | |
| B. siamorientalisKowasupat et al., 2012a, Kowasupat et al., 2012b⁎ | T, Cm & V | C & P | |
T: Thailand, L: Lao PDR, V: Vietnam, B: Brunei, I: Indonesia, M: Malaysia, My: Myanmar, Cm: Cambodia.
Remark: Fighting fish species in Rüber et al. (2004) not included in this study are as follows: B. anabatoides, B. fusca, B. cf. picta, B. breviobesus, B. dimidiata, B. cf. albimarginata “Pampang”, B. cf. albimarginata “Malinau”, B. cf. burdigala, B. brownorum, B. rutilans, B. miniopinna, B. strohi, B. foerschi, and B. simorum.
Fighting fish species in this study that were not included in Rüber et al. (2004).
Geographical origins of the fishes
Fishes in this section are named in accordance with previous publications on the type-locality ones (Kowasupat et al., 2012a, Kowasupat et al., 2012b, Schindler and Schmidt, 2006, Schindler and Schmidt, 2008, Tan and Ng, 2005). A simple map is provided here (Fig. 1) to show Southeast Asian countries where the betta fishes have been reported and some of the places where the type-locality fishes were caught for this work.
Fig. 1.
Map of Southeast Asia showing our collection sites (with symbols) of mouth-brooding betta type-locality species (purple pentagon: B. prima; white pentagon: B. pallida; pink star: B. simplex; white star: B. kuehnei; red triangle: B. apollon; white triangle: B. ferox, yellow diamond: B. stigmosa; blue circle: B. pugnax; white circle: B. pulchra; green hexagon: B. pi; white hexagon: B. waseri; orange square: B. hipposideros and B. livida).
Of the mouth brooders, B. prima fish were found in the eastern provinces of Thailand, in Cambodia and in south Vietnam between latitude 14°N and 9°N. Between 12°N and near 10°N on the mainland Thailand, no brooders have ever been reported. They have been found again only from latitude 9°N in lower peninsular Thailand all the way through mainland Malaysia and some Indonesian islands (6°S). Betta pallida brooders were found in the lower southern provinces of peninsular Thailand where some habitats of Betta ferox and Betta apollon are in close proximity: B. ferox have a wider distribution. Betta pugnax spread from lower Thai Peninsula down to the southern tip of the Malay Peninsula. Betta pi were caught in Thai provinces bordering Malaysia. Betta stigmosa, Betta pulchra, Betta kuehnei, Betta waseri and Betta hipposideros were captured in the localities of type species in peninsular Malaysia. Betta ocellata were found in Sabah on Borneo Island which encompasses the eastern Malaysian states of Sabah and Sarawak, the Kalimantan part of Indonesia, and the country of Brunei. Betta macrostoma were from Brunei. Betta enisae, Betta unimaculata, Betta patoti, Betta ideii, Betta compuncta, Betta krataios and Betta edithae were from the Kalimantan part of the Borneo island in Indonesia whereas Betta rubra and B. chloropharynx were from the Sumatra island, Indonesia.
For the non-Thailand nest builders, the northernmost Betta bellica were found in Malaysia (latitude 4°N) all the way down to southern Malaysia and Sumatra island of Indonesia. Betta coccina group members, e.g., Betta livida, B. coccina, Betta tussyae, Betta rubra, and Betta persephone, were from the Malay Peninsula. B. splendens, B. smaragdina, B. imbellis, Betta stiktos, B. mahachaiensis and Betta siamorientalis were caught and obtained in Thailand and neighboring countries, viz Lao PDR, Myanmar, Cambodia and Vietnam.
Previous classifications
In 2005, Tan and Ng revised the taxonomy of 23 betta fighting fish species in Southeast Asia. Schindler and Schmidt (2006) reported on the reclassification of the six mouth brooders found in Thailand with names given up to that time. Several Southeast Asian betta fishes have been found subsequent to the publication of their works (Kowasupat et al., 2012a, Kowasupat et al., 2012b, Schindler and Linke, 2013, Schindler and Schmidt, 2008, Tan, 2009a, Tan, 2009b, Tan, 2013).
Difficulties in identifying some brooders morphologically
Some mouth brooders have clear body and fin colors and markings, and retain at least some of these characteristics and distinguishing external features regardless of their physiological states, e.g., Betta channoides, B. unimaculata, and B. macrostoma. These are easy to identify morphologically. Other mouth brooders, however, having generally matte, dull and brownish bodies and fins to start with, at times change dark markings and stripes (longitudinal as well as transverse ones) on the bodies causing even the experts to confuse some species with others, e.g., B. ferox, B. pugnax, B. apollon, B. pallida, and B. prima. Much earlier, there had been four cases of misidentification of mouth brooders by Smith (1945), Suvatti, 1950, Suvatti, 1981, Monkolprasit et al. (1997), and Vidthayanon et al. (1997) based on morphology. They misnamed a species of eastern Thailand fish (probably B. prima) as Betta taeniata; the latter are located on the Borneo Island and not in Thailand. Also there was a case of B. prima misidentified as B. taeniata in Vietnam by Anh and Phú (2010). Thus for these fishes the use of DNA sequences should provide a more objective identification alternative to morphological criteria which require more experience in visual observation.
Previous genetic works
A randomly amplified polymorphic DNA (RAPD) analysis of B. prima, B. simplex, B. pugnax, and B. pi from four southern provinces of Thailand was carried out by Tanpitayacoop and Na-Nakorn (2005). A phylogenetic tree of 30 fighting fishes, including B. prima, B. simplex, and B. pugnax, was reconstructed by Rüber et al. (2004) using the complete cytochrome b (cyt b) and tRNA Val genes, and fragments of 12S rRNA and 16S rRNA genes of the mitochondrion, and the nuclear RAG1 gene, all of which were not used in our study. In addition, Lee et al.'s (2012) work on 18S rRNA gene from B. pi showed that the species had a taxon-specific DNA sequence. Also Jamaluddin et al. (2010) had gene-based (16S rRNA gene) evidence for B. pugnax having varieties.
DNA barcoding in fish utilizing DNA of the cytochrome c oxidase subunit I (COI) alone has been shown by several investigators to be helpful and effective in species identification (Pereira et al., 2013, Ward et al., 2009, Zemlak et al., 2009); although in some cases it has been shown to need additional DNA data (Dasmahapatra et al., 2010, Taylor and Harris, 2012). In 2014, Kowasupat et al. reconstructed a phylogenetic tree for the bubble-nesting betta fishes in Thailand by using mitochondrial COI gene and nuclear ITS1 (internal transcribed spacer 1) gene. The ITS1 gene, albeit showing only small differences among species, is still useful in identifying species of the bubble-nesting bettas and is also capable of yielding genetic markers. The combined COI and ITS1 phylogenetic tree shows the relationship of six clearly distinguishable species, viz B. splendens, B. mahachaiensis, B. siamorientalis, B. imbellis, B. smaragdina, and B. stiktos. Close morphological similarities between B. imbellis (in southern Thailand) and B. siamorientalis (in eastern Thailand and Cambodia) had led to doubts about the latter's species status, which was later confirmed by the comparison of COI and ITS1 DNA from the two fishes. Betta smaragdina's cryptic species have also been clearly identified and put in the proper phylogenetic context in the same way.
The above-mentioned DNA sequences and their analyses have thus been useful for genetic species identification and for showing the phylogenetic relationship of the fishes. In our integrative work reported here, the results from using the COI and ITS1 sequences had to be compared with those carried out earlier based on morphology and different DNA sequences (see also Pires and Marinoni, 2010). In this article on Southeast Asian mouth-brooding betta fishes, we show that the species status of certain brooders may have to be revised and also propose a new phylogenetic tree based on COI and ITS1 sequences and their analyses. Some nest builders are also included to make the phylogenetic relationships more complete.
Materials and methods
Sample collection
All fishes in Thailand and some in peninsular Malaysia were caught by us. Those from Cambodia, Vietnam, Brunei, eastern Malaysia (Sabah and Sarawak) and Indonesia (Kalimantan and Sumatra) were supplied to us by experienced traders of the mouth brooders. Some non-Thailand nest-builders were also caught and bought. We usually ordered from two different traders to compare the morphology and DNA sequences of the fishes. The appearances of these fishes generally agreed with photographs and descriptions published previously (Cardot, 2010, Goldstein, 2004, Kottelat et al., 1993, Linke, 2009, Tan and Ng, 2005). The shapes and color patterns on their heads, bodies, and fins, and the rays, bars, and spots on those fins were compared with those from the original publication on each species, as well as the keys to Betta species by Witte and Schmidt (1992), Tan and Ng (2005) and Schindler and Schmidt (2006), in order to identify our specimens.
Thailand mouth-brooding bettas were wild caught from 11 provinces: Chantaburi, Trat and Sa Kaeo in the eastern region, and Krabi, Surat Thani, Nakhon Si Thammarat, Phatthalung, Trang, Satun, Songkla, and Narathiwat in the southern peninsula. Six mouth-brooding species, viz B. simplex, B. prima, B. pallida, B. ferox, B. apollon, and B. pi were collected from the places in Thailand where the type-locality fishes were reported earlier. We found the betta brooders in about 1 to 8 collection sites in each province. One to twenty fishes were randomly taken from each collection site, be it river, stream, waterfall, swamp, peat swamp or pond. Betta prima from Cambodia and southern Vietnam (near Ho Chi Minh City) were purchased. Additional type-locality B. pugnax, B. kuehnei, B. stigmosa, B. waseri, B. hipposideros, B. livida, and Betta tussyae were caught in Malaysia. Betta bellica, B. persephone, and B. ocellata from Malaysia, and B. enisae, B. unimaculata, B. patoti, B. ideii, B. compuncta, B. krataios, B. edithae, B. rubra, B. bellica, B. chloropharynx, and B. coccina from Indonesia were bought for comparison. Betta macrostoma fish were bought.
All fighting fishes caught and bought by us were alive before being anesthetized by putting in ice water at 2–5 °C; the fishes then became unconscious and died quickly. The dead fishes were photographed by using a scanner with a color bar and a ruler scale. The specimens were preserved in 70% ethanol for long-term storage and deposited at Thailand History Museum, Patum Thani (THNHM) and also kept in the authors' own collection for the Thailand Betta Project (TBP). From the right side of each fish specimen muscles were collected, preserved in 95% ethanol and kept at − 20 °C for genetic analysis.
DNA extraction
Approximately five mg of muscle of each fish specimen, randomly selected from available specimens, be it numerous (caught fish) or a few (bought fish), was subjected to DNA extraction using the DNA extraction kit of Stratagene (Agilent Technologies) or following the previous protocol described in Kowasupat et al. (2014). In particular, the tissue was incubated with 180 μl of 50 mM NaOH at 95 °C for 10 min prior to the addition of 20 μl of 1 M Tris− HCl, pH 8.0. The supernatant containing the DNA template was then collected into a new tube to be used in subsequent PCR reactions.
DNA amplification reactions
Two regions analyzed in this study were the barcoding region of the mitochondrial COI gene and the nuclear ITS1 gene. Primers and PCR conditions for COI and ITS1 amplification were employed according to the protocols for the direct sequencing method and DNA cloning method described in a previous work (Kowasupat et al., 2014) with slight modification.
PCR amplification and sequencing of COI
PCR amplification reactions were conducted according to Kowasupat et al. (2014) by using the primers for the mitochondrial COI gene including VF2_t1, FishF2_t1, FishR2_t1, and FR1d_t1 as previously described (Ivanova et al., 2007, Ward et al., 2005). In direct sequencing, PCR products were obtained using M13F(− 20) and M13R-pUC(− 26) as primers (see Table 2).
Table 2.
Primers used in PCR amplification and DNA sequencing of the COI and ITS1 regions.
| Usage | Primer name | Primer sequence (5′–3′) | References |
|---|---|---|---|
| COI | VF2_t1⁎ | TGTAAAACGACGGCCAGTCAACCAACCACAAAGACATTGGCAC | Ward et al. (2005) |
| COI | FishF2_t1⁎ | TGTAAAACGACGGCCAGTCGACCTAATCATAAAGATATCGGCAC | Ward et al. (2005) |
| COI | FishR2_t1⁎ | CAGGAAACAGCTATGACACTTCAGGGTGACCGAAGAATCAGAA | Ward et al. (2005) |
| COI | FR1d_t1⁎ | CAGGAAACAGCTATGACACCTCAGGGTGTCCGAARAAYCARAA | Ivanova et al. (2007) |
| ITS1 | Betta_ITS1_F1 | CACACCGCCCGTCGCTACTA | Kowasupat et al. (2014) |
| ITS1 | Betta_ITS1_F2 | ACTTGACTATCTAGAGGAAG | Kowasupat et al. (2014) |
| ITS1 | Betta_ITS1_R1 | GTYCTTCMTCGACSCACGAG | Kowasupat et al. (2014) |
| ITS1 | Betta_ITS1_R2 | GTTCTTCATCGACGCACGAG | Kowasupat et al. (2014) |
| ITS1 | Betta_ITS1_R4 | TCCACCGCTAAGAGTTGTC | Kowasupat et al. (2014) |
| Sequencing | M13F(− 20) | GTAAAACGACGGCCAGT | Messing (1983) |
| Sequencing | M13R-pUC(− 26) | GGAAACAGCTATGACCATG | Messing (1983) |
| Sequencing | M13R(− 24) | CAGGAAACAGCTATGAC | Messing (1983) |
| Sequencing | SP6 | ATTTAGGTGACACTATAG | – |
| Sequencing | T7 | AACAGCTATGACCATG | – |
In DNA cloning method for COI, leading M13 sequences (underlined) were omitted from the primers indicated.
For the samples using the DNA cloning method, the primers used were as above without the leading sequences of M13 as in Table 2. After agarose gel analysis and DNA purification, the gel-purified fragments of about 700 base pairs (bp) were then ligated to the pPrime cloning vector (5PRIME). The ligation mixture was then transformed into Escherichia coli XL-1Blue cells and the transformants were plated onto LB-agar containing 50 μg/ml ampicillin. Recombinant clones with correct insert size verified by colony PCR were subjected to plasmid extraction using Plasmid DNA Extraction Mini Kit (Favorgen Biotech Corp.). The recombinant plasmids were then sequenced. All fragments were fully sequenced from both strands of the DNA using vector primers, i.e., SP6 and T7 promoter primers (see Table 2).
All samples were sequenced by 1st BASE DNA Sequencing Services (Malaysia).
PCR amplification and sequencing of ITS1
ITS1 PCR reactions and conditions followed those previously described by Kowasupat et al. (2014) with a minor change. Most PCR products used Betta_ITS1_F2 and Betta_ITS1_R4 primers (as shown in Table 2) for amplification of the nuclear ITS1 gene. These primers were synthesized by Bio Basic Inc., Canada. The universal M13F(− 20) and M13R(− 24) sequencing primers were included in the 5′-end of the former and the latter amplification primers respectively to enable direct sequencing. After agarose gel electrophoresis, amplified products of approximately 500 bp were subjected to gel purification using Gel/PCR Purification Kit (Favorgen Biotech Corp.). Direct sequencing using universal M13F(− 20) and M13R(− 24) primers was then carried out by 1st BASE DNA Sequencing Services (Malaysia). The remaining samples used primers Betta_ITS1_F1 paired with Betta_ITS1_R1 or Betta_ITS1_F1 paired with Betta_ITS1_R2 for amplification and sequencing as described in a previously established protocol (Kowasupat et al., 2014). These primers were synthesized by Sigma-Aldrich Co. LLC., USA. DNA sequencing was performed by 1st BASE DNA Sequencing Services (Malaysia).
Sequence submission
Fighting fish sequence data were submitted to GenBank (www.ncbi.nlm.nih.gov). Of the total 707 COI sequences, 306 were from Kowasupat et al., 2014, 249 of the bubble-nest builders were from Sriwattanarothai et al., 2010, plus 152 new COI sequences of the mouth brooders with accession numbers KM485311 to KM485462. The percentage of base-pair differences from the total of 652 bp in the COI sequence was used for intra-specific and inter-specific sequence comparison. Of the total 192 ITS1 sequences, 102 were from Kowasupat et al. (2014). All the new 90 ITS1 sequences in this study have accession numbers KM485463 to KM485552.
Reconstruction of phylogenetic tree
To ensure the integrity of the phylogenetic analyses, the researcher who performed the analyses did not know until the end of tree building from which specimens (all coded by numbers) the DNA sequences were extracted, and at the end no changes were made to the final tree.
Sequence alignment
For specimens whose forward and reverse DNA sequencing results were available, the sequences were assembled using Geneious version 5.5.9 (Biomatters Limited; www.geneious.com). If the assembling failed, the sequences were aligned instead. The assembly or the alignment was then visually inspected to ensure that the forward and reverse sequences agreed. For each specimen, the required region of the sequence was then extracted.
The extracted sequences together with the consensus sequences of all the species/cryptic species in the B. splendens and B. smaragdina groups (Kowasupat et al., 2014, Sriwattanarothai et al., 2010) were aligned using MUSCLE (Edgar, 2004) that was available in Geneious with the option of grouping similar sequences together so that the most likely sequences of multi-sequence specimens could be selected by experienced fish experts (not by the one who performed the alignment). The alignments were then inspected and adjusted manually. The adjustment to the COI alignment was minimal. Besides MUSCLE, Geneious and ClustalW (Thompson et al., 1994) were also used to align the ITS1 sequences, all with several sets of options. The resulting alignments were quite different, with the alignment lengths ranging from about 500 to 650 bp, and all of them seemed unsatisfactory. This was probably due to the fact that the ITS1 region had gone through many insertions and deletions over time, resulting in the alignment that contained one huge indel region and several small ones. One of the ITS1 alignments was modified so that the grouping of the specimens in the resulting phylogenetic tree would be as neutral as possible while keeping the gaps moderate.
Phylogenetic analysis
In addition to the COI and ITS1 datasets, a combined dataset was created by joining the two DNA regions of the same specimen. All repeating sequences in the each dataset were eliminated so that the sequence of each specimen was unique. Phylogenetic analyses were performed on the three datasets using MrBayes version 3.2.2 (Huelsenbeck and Ronquist, 2001, Ronquist and Huelsenbeck, 2003, Ronquist et al., 2012). First, preliminary Bayesian inference was carried out for each of the dataset using the most extensive single-nucleotide model for that dataset. Metropolis-coupled Markov chain Monte Carlo (MC3) was used to sample from the posterior distribution in order to observe the characteristics of the run using Tracer version 1.5 (Rambaut and Drummond, 2003). The posterior distribution was used to decide which evolutionary models, and in which order, should be inspected for their marginal likelihoods so that their fitness for the given dataset could be compared using Bayes factors. The marginal likelihoods (ML) were then estimated using the stepping-stone (SS) method proposed by Xie et al. (2011) and implemented in MrBayes version 3.2 and subsequent versions. Before SS was available, ML was usually estimated using the harmonic mean (of the posterior likelihoods) which was reported by MrBayes after an MC3 run and often greatly overestimated true ML (see, for example, Fan et al., 2011, Xie et al., 2011). We thus adopted the SS method with the aim of improving the results. The model with the lowest ML estimate would be selected for final Bayesian inference.
Results
Sequence alignment
There were 93 unique COI sequences, 74 unique ITS1 sequences, and 83 unique combined sequences (including 10 consensus sequences from the B. splendens and B. smaragdina groups). The alignment of COI involved 652 bp while that of ITS1 was 502 bp. The COI alignment contained 299 variable sites, 267 of which were informative. Of the informative sites, 51, 10, and 206 were from the first, second, and third positions of the codons respectively. Differences in base pairs of DNA sequences from each specimen were compared and analyzed.
New findings on species status
Before presenting our findings, a few words about posterior probabilities are in order. The posterior probability of a clade is the probability that, given the data (the sequence alignment), all the taxa in that clade have a recent common ancestor which is not the ancestor of any taxon outside the clade. Thus, a posterior probability close to 1 (say, > 0.9) is a good indication of the speciation of that clade while a low posterior probability indicates (< 0.8) that the grouping may happen by chance or that there are other grouping alternatives.
Our findings cast new light on four species currently recognized as being different. The Betta prima and B. pallida pair seemed to be genetically indistinguishable as the posterior probabilities of the two clades were only 0.36 and 0.79 respectively in the COI tree and the specimens of B. pallida could not even form a clade in the combined tree (Fig. 2, Fig. 3). Moreover, the COI sequences of the two differed by only 5.87 bp (0.9%) on average. Even though B. prima seemed to be genetically indistinguishable from B. pallida, 5 specimens of the former formed a subgroup within this clade (B. sp. (cf. prima)) with posterior probability of 0.98 in the combined tree. The average difference among the COI sequences in this subgroup was 2.5 bp (0.38%) while the sequences differed from those in the main group by 15.5 bp (2.38%) on average. The COI genes of the other two brooders, B. apollon and B. ferox, were so nearly identical that the specimens of B. apollon could not form a monophyletic clade. Nonetheless, 3 subsets of the former formed 3 monophyletic clades, one (type-locality B. apollon) with posterior probability of 1 in the combined and COI trees, the other two (B. spp. (cf. apollon) 1 and 2) with posterior probability of 1 in one of the two trees and more than 0.9 in the other tree. The COI sequences within each B. apollon subgroup differed by only about 1 bp on average while the average differences among these subgroups were 8.5–9 bp (1.30–1.38%). Moreover, two specimens of B. ferox also formed a subgroup (B. sp. (cf. ferox)) with posterior probability of 1 in the combined and COI trees. The COI sequences of the two specimens in this subgroup were identical while they differed from those of other B. ferox (type-locality) by 6.5 bp (1%). Contrary to the two cases above, our results suggested the possibility of a new cryptic species or even a new species of B. simplex currently known as a single species. B. simplex specimens formed 2 monophyletic clades (type-locality B. simplex and B. sp. (cf. simplex)) with the posterior probability of 1 (combined and COI trees) for both clades. The COI sequences of the two clades differed by 34.17 bp (5.24%) on average while the differences within the clades were only 1 and 2 bp (0.15% and 0.31%).
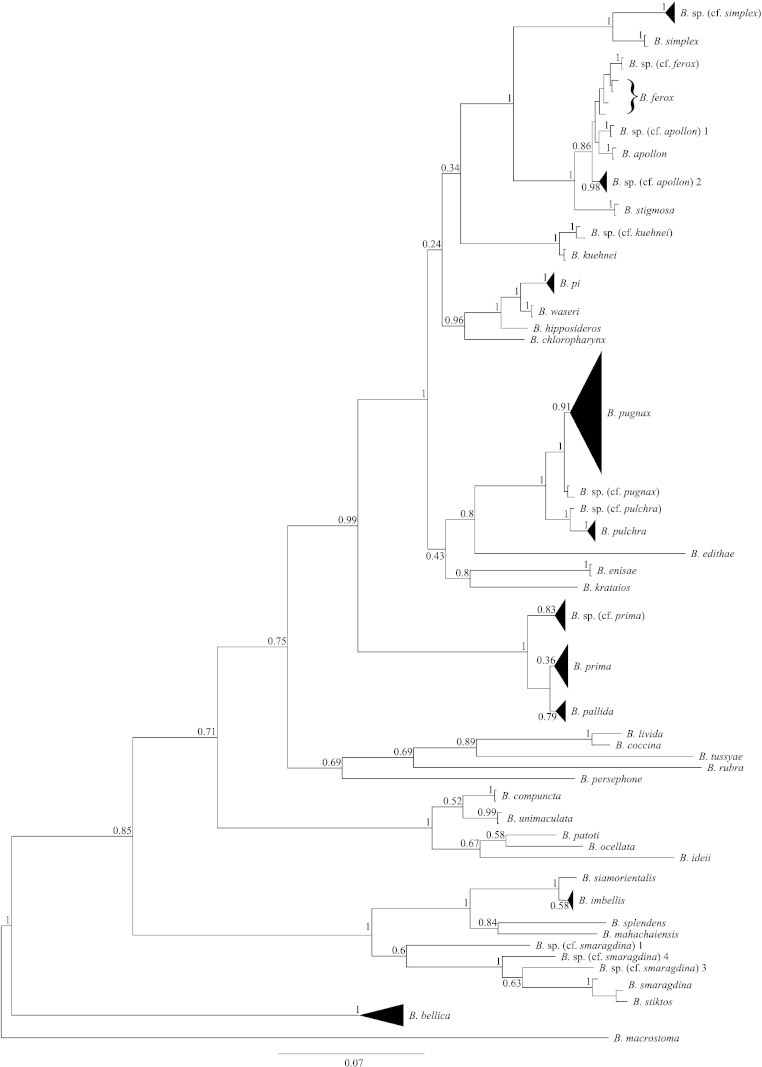
Fig. 2.
The phylogenetic tree reconstructed from the alignment of 83 COI and ITS1 sequences using Bayesian inference with Metropolis-coupled Markov chain Monte Carlo (MC3): the posterior probabilities are indicated next to the nodes. The dataset was divided into 4 partitions: one for each codon position and another for ITS1. An isosceles triangle 's depth (along the tree) corresponds to the depth of the most divergent specimen in the clade while its width represents the number of varieties. Each terminating line represents a single specimen.
Fig. 3.
The phylogenetic tree reconstructed from the alignment of 93 COI sequences using Bayesian inference with Metropolis-coupled Markov chain Monte Carlo (MC3): the posterior probabilities are indicated next to the nodes. The dataset was divided into 3 partitions according to the positions in the codons. An isosceles triangle's depth (along the tree) corresponds to the depth of the most divergent specimen in the clade while its width represents the number of varieties. Each terminating line represents a single specimen.
Phylogenetic relationship
The phylogenetic trees inferred from the three datasets (COI, ITS1, and COI + ITS1) are shown in Fig. 2, Fig. 3, Fig. 4. A preliminary inference from the COI (652 bp) dataset using Trichopsis vittata as the outgroup revealed that B. macrostoma should be the root of the trees (result not shown) which was also the conclusion of Rüber et al. (2004). In addition, Table 3 shows that the COI sequence of B. macrostoma differed the most from those of other specimens. Thus, B. macrostoma was used to root the three inferred trees. Generally, the phylogenetic relationships among the taxa were similar to those in Rüber et al. (2004). As indicated by the inferred combined and COI trees, B. macrostoma should not be a member of B. unimaculata group. In Rüber et al., even though the two taxa speciated from other taxa consecutively, the former could not form a clade. Besides B. macrostoma and B. unimaculata group, B. bellica, B. splendens group, and B. smaragdina group seemed to speciate from the other fishes very early on. The grouping of Betta foerschi group (represented by B. rubra in our COI tree) and B. coccina group was also supported albeit very weakly with posterior probability of 0.69 (COI tree only). In Rüber et al. (2004), B. simplex formed a clade with B. cf. picta and the clade, in turn, was a sister group to the clade containing B. pugnax. Since neither the sequence of B. picta nor that of B. cf. picta was obtained, B. simplex was a sister group to the clade containing B. apollon and B. ferox (not present in Rüber et al.), members of B. pugnax group, instead. In our COI tree and that of Rüber et al., B. edithae should not be a member of B. picta group as indicated by Schindler and Schmidt (2006).
Fig. 4.
The phylogenetic tree reconstructed from the alignment of 74 ITS1 sequences using Bayesian inference with Metropolis-coupled Markov chain Monte Carlo (MC3): the posterior probabilities are indicated next to the nodes. An isosceles triangle's depth (along the tree) corresponds to the depth of the most divergent specimen in the clade while its width represents the number of varieties. Each terminating line represents a single specimen.
Table 3.
The average of all the pairwise differences between the COI sequence(s) of a clade and those of the remaining taxa in the inferred COI tree (see Fig. 3). The word “group” (used only in this table) reflects the clustering in our tree, not the species groups of Witte and Schmidt (1992), Tan and Ng (2005), or Schindler and Schmidt (2006).
| Clade | Difference (bp) | Difference (%)⁎ |
|---|---|---|
| B. macrostoma | 144.36 | 22.14 |
| B. bellica | 130.21 | 19.97 |
| B. splendens group & B. smaragdina group | 126.23 | 19.36 |
| B. rubra & B. coccina group | 115.24 | 17.67 |
| B. unimaculata group | 111.54 | 17.11 |
| B. prima group | 101.80 | 15.61 |
| B. edithae | 100.09 | 15.35 |
| B. simplex group | 96.78 | 14.84 |
| B. pugnax group | 93.38 | 14.32 |
| B. stigmosa & B. apollon group | 93.29 | 14.31 |
| B. enisae | 91.64 | 14.05 |
| B. krataios | 90.20 | 13.83 |
| B. kuehnei group | 89.12 | 13.67 |
| B. waseri group | 83.92 | 12.87 |
From the total of 652 base pairs.
Some phylogenetic relationships in our results are notably different from those in the literature. Betta waseri group formed a clade with B. prima and B. pallida with posterior probability of 1 in both the combined and ITS1 trees (500 bp) while the former was phylogenetically closer to B. pugnax and B. simplex in Rüber et al., 2004. Whereas their results suggested that the speciation of B. unimaculata group happened before that of B. bellica, B. splendens, and B. smaragdina groups, all our trees indicated that the reverse was true.
The majority of B. pugnax formed a monophyletic clade with posterior probability of 1 in the combined tree. The remaining two specimens formed another clade (B. sp. (cf. pugnax)) with very low posterior probability in all our trees (~ 0.6). The COI sequences in the latter clade differed by 2 bp (0.31%) while those of the former clade differed by 4.02 bp (0.62%) on average. The sequences of the two clades differed by 9.08 bp (1.39%) on average. All except one specimen of B. pulchra formed a monophyletic clade with posterior probability of 1 in the combined and COI trees. The COI sequences within this clade differed by 2.67 bp (0.41%) on average while they differed from that of the other specimen (B. sp. (cf. pulchra)) by 8.33 bp (1.28%) on average. The specimens of B. kuehnei also formed 2 monophyletic clades with posterior probability of 1 in the combined and COI trees for one of the clades (B. sp. (cf. kuehnei)). Due to the low posterior probability (< 0.7 in both trees) of the other clade (type-locality B. kuehnei), the former might be a subgroup instead of an independent group. The COI sequences of the two specimens within this subgroup differed by 4 bp (0.61%) while they differed from the sequences of other B. kuehnei specimens by 9 bp (1.38%) on average.
The preceding results on species and their phylogenetic relationships were made possible and more credible by the results below.
Phylogenetic analysis
The aforementioned phylogenetic relationships were obtained after following the procedures described in section 2.5.2. After many SS runs, the 2-parameter nucleotide substitution model was selected for the second COI codon partition while the reversible-jump (mixed) nucleotide substitution model (Huelsenbeck et al., 2004) was selected for the third COI codon and the ITS1 partitions. Interestingly, the model choices for the first COI codon partitions of the combined and COI datasets were different: the choice for the combined dataset was the general time-reversible model while that for the COI dataset was the mixed model. Regarding the choices for rate variations across sites, + G + I was best for the second COI codon partition while + G was best for the third COI codon and the ITS1 partitions. Once again, the choices for the first COI codon partitions of the two datasets were different: + G for the combined dataset and + G + I for the COI dataset. The inconsistency of the model choices for the first COI codon partitions of the two datasets indicated that each Bayesian analysis had to be performed independently: one should not employ the model choice from a dataset in the analysis of another dataset.
Discussion
Species status
Although the overall phylogenetic tree presented herein generally agrees with the one in Rüber et al. constructed in 2004, some species and groups are not positioned on the tree as in their report. Also using COI and ITS1 gene sequences and analyses has led to results on species that do not always in accord with previous assignments.
The B. pallida (southern Thailand) and B. prima (eastern Thailand) pair long suspected as being morphologically nearly identical, could now be considered to be genetically identical and thus could be merged into a single species. Naturally, more loci will help to validate the result.
The southern B. ferox and B. apollon which appear very similar now have been shown to be very close genetically by COI and ITS1 sequences and analyses to the extent that they could be considered identical, therefore forming possibly one single species. As stated above more work is needed to prove the identity.
One finding, totally unexpected from similarities in body shape and fin color, is that B. simplex (endemic to southern Thailand) in Krabi (type locality being Tham Sa Kaeo as in Kottelat, 1994) from two different habitats not too far apart (about 50 km) are genetically different enough to be considered different species. The genetic evidence for a new species of B. simplex was very strong; the COI sequences of the two subgroups differed by more than 10 times the difference within each subgroup and the posterior probabilities for both clades were 1 in the combined and COI trees. However, upon careful inspection of fish specimens (N = 25) from the two catch sites the caudal transverse bars could distinguish the great majority of the type-locality fish (mostly absent) from that of the other fish (mostly present) caught at the other site. Realizing possible controversies raised by our results above, we had made several trips to catch the fishes and repeated our DNA experiments to convince ourselves of the validity.
Some small gradations in DNA sequence have been found in specimens of established species. Much larger differences though may lead to possible cryptic species. There is a potentially new cryptic B. prima species from south Vietnam (B. sp. (cf. prima)) with COI sequences differing from those of specimens with the same name from Thailand by more than 10 bp on average; the posterior probability of this clade was 0.97 in the combined tree, a relatively high value. Thus, these south Vietnamese B. prima are genetically different enough from Thailand B. prima to be treated as at least a cryptic species. Vietnamese B. prima have transverse bars on the caudal fin whereas Thailand B. prima and those obtained from Cambodia do not have this feature. Although there were other potentially new cryptic species with high posterior probabilities, their COI sequences were less than 10 bp different from those of others in the same species. Thus, their cryptic-species statuses were not as strongly supported.
There are two clades, namely B. apollon–B. ferox–B. stigmosa clade and B. pugnax–B. pulchra clade, whose DNA results indicate gradations of sequence differences in the populations from habitats in southern Thailand all the way down to lower peninsular Malaysia. As regards the first clade, B. sp. (cf. ferox), B. ferox, B. sp. (cf. apollon) 1, B. apollon, B. sp. (cf. apollon) 2, and B. stigmosa show a gradation of COI sequences among the taxa which corresponds to their distribution from southern Thailand to the northern part of Malaysia. However, there is no observable geographical pattern of gradation within any taxon. Concerning the second clade, a similar gradation ranges from B. pugnax in southern Thailand to B. sp. (cf. pugnax), B. sp. (cf. pulchra), and B. pulchra at the tip of Malay Peninsula. Geographical gradation of habitats can also be observed among the members of B. waseri group, namely B. pi, B. waseri, B. hipposideros, and B. chloropharynx, from north to south respectively.
In the literature (Hebert et al., 2004, Mueller, 2006), the threshold of 10 for the ratio of inter-clade to intra-clade differences in COI sequences has been proposed to decide whether the clades are two distinct species. This threshold number seems to be too conservative. The aforementioned ratios for several pairs of well-established species in this study are about 5 (e.g. B. waseri–B. pi and B. stigmosa–B. ferox, data not shown). In fact, these ratios should be reflected in the posterior probabilities of these clades provided that a sufficient proportion of all the varieties of each species is sequenced. The same can be said for the threshold of 2% for the inter-clade difference. Thus, as long as the posterior probability supporting a clade was close to 1, this clade could be considered a potential new species or cryptic species (designated as cf. species, i.e., B. sp. (cf. prima), B. sp. (cf. simplex), B. spp. (cf. apollon) 1 and 2, B. sp. (cf. ferox), B. sp. (cf. pugnax), B. sp. (cf. pulchra), and B. sp. (cf. kuehnei)).
Although the discordance in the parental origin of COI and that of ITS1 obtained from the same specimens would indicate existence of hybrids, we have not found any hybrid fish although there were places where more than one species of brooder cohabited. We have been able to interbreed B. kuehnei and B. pugnax to produce live fry. Thus if hybrids did exist in the fishes studied, we would probably have found them.
New phylogenetic tree
Consulting Tan and Ng's (2005) table on members of the betta fishes according to their being bubble nesters or mouth brooders and their previous assignments as to which main group each member belonged to, one can see that our reconstructed tree agrees with their group assignments in general. For example, members of the brooder B. waseri group (B. chloropharynx, B. hipposideros, B. pi, and B. waseri) previously put together because of some clear morphological similarities remain together on our tree. Members of bubble nesters in the B. coccina group (B. coccina, B. livida, and B. tussyae) also stay together as before (Rüber et al., 2004).
By extension, it is also to be expected that members of the bubble nesters should be grouped together and the same applies to members of the mouth brooders because of such a clear-cut behavioral difference (Schindler and Schmidt, 2006, Tan and Ng, 2005, Witte and Schmidt, 1992).
However, our results (based on mitochondrial COI and nuclear ITS1) also are not all in accord with Tan and Ng's (2005) and Schindler and Schmidt's (2006) earlier groupings (based on morphology) in a few aspects, especially B. pugnax group and B. picta group.
Our COI and ITS1-based tree has features similar to those of Rüber et al. (2004) with some differences. That the posterior probability of the B. simplex–B. apollon clade in the combined and COI trees being 1 did not necessarily contradict the results in Rüber et al. (2004) that grouped B. simplex with B. cf. picta as long as B. edithae and B. enisae did not belong to B. picta group. In fact, Rüber et al. also found that B. edithae was not a member of B. picta group. It seems as if close relatives of either B. edithae or B. enisae have yet to be found or their DNA sequences have yet to be analyzed.
In the COI tree, B. waseri group is sister to B. kuehnei group with extremely low posterior probability of 0.33 while the former is sister group to B. prima and B. pallida in the ITS1 tree with posterior probability of 1. The latter is an example of the inability of COI in determining the phylogenetic relationship of distantly related species. Although the position of B. waseri group in Rüber et al. appears more similar to its position in the COI tree than that in the ITS1 tree, we believe that the ITS1 one is more credible. Rüber et al. used several mitochondrial genes and a protein-coding nuclear gene to reconstruct their tree. The combined length of their genes was about 4 times the length of our combined sequence. Yet cyt b evolves at a rate similar to COI while the mitochondrial ribosomal genes and RAG1 might evolve too slowly to be able to determine some of the phylogenetic relationships among groups of species in the same genus.
Although differences between sequences of the two genes may not always reflect obvious morphological differences and vice versa, our species groupings from the two genes are in general supportive of obvious morphological similarities. The results on the cases of morphologically nearly identical pairs, viz B. prima vs B. pallida and B. ferox vs B. apollon further confirm the usefulness of employing the two genes to help decide species status.
We did not utilize the RAG1 gene because from our experience it did not change the COI&ITS1 combined tree much. Also there is a publication (Rüber et al., 2006) stating that RAG1 is not so useful for distinguishing at the species level, being more suitable for higher levels of taxon.
Ancestry and evolution
The eastern Thailand B. prima fish are identical to the B. pallida in terms of DNA sequences (mitochondrial COI and nuclear ITS1) even though the habitats of the two populations have been isolated by the rising water level in the Gulf of Thailand since the end of the last ice-age (~ 20,000 years) (Voris, 2000, Woodruff, 2010), presumably not long enough separation time to make the two DNA genes to differ sufficiently. In fact these brooder fishes are less different than the nester pair presently inhabiting the same two distinct regions, viz B. siamorientalis (eastern Thailand) and B. imbellis (southern Thailand), which apparently have evolved more far apart since the submersion (due to molten ice) of the land bridges (with interconnecting river systems and water bodies) formerly linking their present distant habitats. It is noted that B. pallida populations are found in provinces facing the Gulf of Thailand and in some of the provinces facing the Andaman Sea.
That B. macrostoma, a brooder, should be ancestral to B. bellica (a nester with similar body shape) and subsequently gives rise to B. splendens / B. smaragdina group (colorful nesters) and later still to many mouth-brooder groups and nest-builder groups should not be too surprising. Earlier Rüber et al. (2004) had studied many genera of fishes and found no evidence that one type of paternal care should precede the other and vice versa. It is the case here that there was more than one switch in the type of paternal care through successive populations.
The evolutionary hot spots may not have been on the landmasses of the present Borneo Island and/or other Southeast Asian region only (de Bruyn et al., 2014) but could possibly also be on the landmass of the so-called Sundaland which emerged from the sea once the sea level was lowered due to sea ice formations. Migrations and/or emigrations could disseminate populations that could become distinct from one another given time and conditions. When the water level rose after the melting of land ice and sea ice separating the populations, fishes began to evolve differently. From the overall phylogenetic tree we should be able to estimate the sequence of evolutionary events from one ancestor to later generations in a way similar to that estimated in Rüber et al. (2006).
Rationales for our sequence alignment and phylogenetic analysis in this study
Gene selection
Generally, to get a reliable inference of the phylogenetic relationships among specimens, several genes, both the nuclear and mitochondrial ones, should be sequenced and analyzed. However, the complexity of phylogenetic inference increases exponentially with the number of taxa and the combined length of these sequences. In this paper, more than 150 specimens were considered. Thus, the combined length and hence the number of genes had to be as low as possible: ideally one short region each from a nuclear gene and a mitochondrial gene. For the latter, this DNA barcoding region was selected because it was short yet powerful in species identification, implying that the region evolved relatively fast compared to other regions in the mitochondrial genome. As such, it should be able to reveal the phylogenetic relationships among closely related species but should not be as good at revealing the phylogenetic relationships among groups of species. (This can be seen from the fact that most of the posterior probabilities of the clades containing two or more groups in the COI tree were significantly lower than 1.) The nuclear region selected for the analysis should be able to bridge this gap. Any coding region of the nuclear genome would evolve too slowly and thus a rather long piece of DNA would be required. So, the non-coding ITS1 gene, which is also short, was selected for the analysis. Since ITS1 contains many indel regions, aligning ITS1 sequences was a challenge. Even though the knowledge about the origins of these sequences should help simplify the alignment, it might also influence the alignment. To prevent this bias, the origins of all the sequences were kept from the researcher who performed the analysis until the very end process.
Phylogenetic analysis
A model that was more suitable for the data should yield a more reliable phylogeny. Thus, the steps in employing SS were carefully designed and the parameters chosen after some calculation and experimentation. The log files of an SS run were inspected to ensure that the sample of log likelihoods was taken after the convergence had been achieved and that the sample size was large enough in each step. From these inspections and the inspections of the resulting ML estimates, our choice of parameters for SS was rather efficient at achieving the required accuracy. Fan et al. (2011) proposed a generalized SS method which was more efficient than the one implemented in MrBayes. Unfortunately, it could be employed only when the topology was fixed.
Conclusions
Based on COI and ITS1 sequences of betta fishes we have found (a) differently named fishes being identical, (b) the possibility of new and cryptic species arising from what hitherto believed to be single betta species, implying both deflation and inflation of species, as well as (c) new ancestral relationships on the phylogenetic tree of these fishes.
The issues arising from above should be settled by experts (on morphology, molecular genetics, informatics, and taxonomy) collaborating in renaming old species and naming new ones. These diverse people should soon be able to put the fishes into species groups and ancestral relationships in light of our findings. The possibility of larger numbers and longer sequences from cheaper, more rapid high through-put sequencing in the near future offers a good prospect to settle these issues soon.
Acknowledgments
This work would not have been possible without the help of such aquarist specialists , A. Phumchoosri, J. Attavichitchanyarak, and H. Linke. We appreciate K. Pinkaew, N. Buafai, A. Nuallaong, H. Saelim, and K. Tongpijit for their laboratory assistance. We are indebted to T. Jeenthong, Dr. B. Nuangsaeng, Dr. N. Sriwattanarothai, Dr. A. Monvises, S. Somadee, and N. Jairuksa for their assistance in sample collections. We thank A. Blanc, R. Blanc, Y. Chaisuntornkitti, P. Soisook, and W. Pipatmekin for providing us with some mouth-brooding bettas, N. Panitvong for giving two B. pugnax and T. Utapong for one B. prima in Thailand, and U.T. Nhản, T. Vịnh, and Z. Pinitpolnikorn for some B. prima from Vietnam. We are also indebted to several other people, not mentioned here, who helped us at betta collecting sites. We thank the Office of Higher Education Commission (National University Research Grant allocated to Mahidol University) which also financially supported the PhD student of C. Kowasupat.
Contributor Information
Bhinyo Panijpan, Email: [email protected].
Chanon Kowasupat, Email: [email protected].
Parames Laosinchai, Email: [email protected].
Pintip Ruenwongsa, Email: [email protected].
Amornrat Phongdara, Email: [email protected].
Saengchan Senapin, Email: [email protected].
Warapond Wanna, Email: [email protected].
Kornsunee Phiwsaiya, Email: [email protected].
Jens Kühne, Email: [email protected].
Frédéric Fasquel, Email: [email protected].
References
- Anh V.T.P., Phú V.V. Dẫn liệu về thành phần loài cá ỡ hệ thống song Thu Bồn — Vu Gia, tỉnh Quảng Nam. Tạp Chí Sinh Học (J. Biol.) 2010;32:12–20. [Google Scholar]
- Cardot J. CIL-IBSC; France: 2010. Illustrated to the genus Betta. [Google Scholar]
- Dasmahapatra K.K., Elias M., Hill R.I., Hoffman J.I., Mallet J. Mitochondrial DNA barcoding detects some species that are real, and some that are not. Mol. Ecol. Resour. 2010;10:264–273. doi: 10.1111/j.1755-0998.2009.02763.x. [DOI] [PubMed] [Google Scholar]
- de Bruyn M., Stelbrink B., Morley R.J., Hall R., Carvalho G.R., Cannon C.H., van den Bergh G., Meijaad E., Metcalfe I., Boitani L., Maiorano L., Shoup R., von Rintelen T. Borneo and Indochina are major evolutionary hotspots for Southeast Asian biodiversity. Syst. Biol. 2014;63:879–901. doi: 10.1093/sysbio/syu047. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Wu R., Chen M.H., Kuo L., Lewis P.O. Choosing among partition models in Bayesian phylogenetics. Mol. Biol. Evol. 2011;28:523–532. doi: 10.1093/molbev/msq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein R.J. Barron's Educational Series, Inc.; New York: 2004. The betta handbook. [Google Scholar]
- Hebert P.D.N., Stoeckle M.Y., Zemlak T.S., Francis C.M. Identification of birds through DNA barcodes. PLoS Biol. 2004;2:1657–1663. doi: 10.1371/journal.pbio.0020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J.P., Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P., Larget B., Alfaro M.E. Bayesian phylogenetic model selection using reversible jump Markov chain Monte Carlo. Mol. Biol. Evol. 2004;21:1123–1133. doi: 10.1093/molbev/msh123. [DOI] [PubMed] [Google Scholar]
- Ivanova N.V., Zemlak T.S., Hanner R.H., Hebert P.D.N. Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Notes. 2007;7:544–548. [Google Scholar]
- Jamaluddin J.A.F., Sharifuddin N., Jian G.T., Yeong L.T., Nor S.A.M. Proceedings of the 7th Regional IMT-GT UNINET and the 3rd Joint International PSU-UNS Conferences. 2010. Systematics and phylogenetic relationships of the Betta pugnax group in Malaysia; pp. 131–133. [Google Scholar]
- Kottelat M. Diagnoses of two new species of fighting fishes from Thailand and Cambodia (Teleostei: Belontiidae) Ichthyol. Explor. Freshwat. 1994;5:297–304. [Google Scholar]
- Kottelat M., Whitten A.J., Kartikasari S.N., Wirjoatmodjo S. Periplus Editions; Jakarta: 1993. Freshwater fishes of western Indonesia and Sulawesi. [Google Scholar]
- Kowasupat C., Panijpan B., Ruenwongsa P., Sriwattanarothai N. Betta mahachaiensis, a new species of bubble-nesting fighting fish (Teleostei: Osphronemidae) from Samut Sakhon Province, Thailand. Zootaxa. 2012;3522:49–60. [Google Scholar]
- Kowasupat C., Panijpan B., Ruenwongsa P., Jeenthong T. Betta siamorientalis, a new species of bubble-nest building fighting fish (Teleostei: Osphronemidae) from eastern Thailand. Vertebr. Zool. 2012;62:387–397. [Google Scholar]
- Kowasupat C., Panijpan B., Laosinchai P., Ruenwongsa P., Phongdara A., Wanna W., Senapin S., Phiwsaiya K. Biodiversity of the Betta smaragdina (Teleostei: Perciformes) in the northeast region of Thailand as determined by mitochondrial COI and nuclear ITS1 gene sequences. Meta Gene. 2014;2:83–95. doi: 10.1016/j.mgene.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.K.Y., Ng S.Y., Yong D., Chai X.C., Yin W.F., Chan K.G. Molecular phylogeny characterization of Malaysian fighting fish Betta pi (Teleostei: Osphronemidae) based on its 18S ribosomal DNA sequences. Asia Life Sci. 2012;21:57–64. [Google Scholar]
- Linke H. European Anabantoid Club; 2009. Betta news special edition 2009. [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Monkolprasit S., Sontirat S., Vimollohakarn S., Sonsirikul T. Office of Environmental Policy and Planning; Bangkok: 1997. Checklist of fishes in Thailand. [Google Scholar]
- Mueller R.L. Evolutionary rates, divergence dates, and the performance of mitochondrial genes in Bayesian phylogenetic analysis. Syst. Biol. 2006;55:289–300. doi: 10.1080/10635150500541672. [DOI] [PubMed] [Google Scholar]
- Pereira L.H., Hanner R., Foresti F., Oliveira C. Can DNA barcoding accurately discriminate megadiverse Neotropical freshwater fish fauna? BMC Genet. 2013;14:20. doi: 10.1186/1471-2156-14-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires A.C., Marinoni L. DNA barcoding and traditional taxonomy unified through integrative taxonomy: a view that challenges the debate questioning both methodologies. Biota Neotrop. 2010;10:339–346. [Google Scholar]
- Rambaut A., Drummond A. University of Oxford; Oxford: 2003. Tracer: MCMC Trace Analysis Tool. [Google Scholar]
- Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüber L., Britz R., Tan H.H., Ng P.K.L., Zardoya R. Evolution of mouthbrooding and life-history correlates in the fighting fish genus Betta. Evolution. 2004;58:799–813. doi: 10.1111/j.0014-3820.2004.tb00413.x. [DOI] [PubMed] [Google Scholar]
- Rüber L., Britz R., Zardoya R. Molecular phylogenetics and evolutionary diversification of labyrinth fishes (Perciformes: Anabantoidei) Syst. Biol. 2006;55:374–397. doi: 10.1080/10635150500541664. [DOI] [PubMed] [Google Scholar]
- Schindler I., Linke H. Betta hendra — a new species of fighting fish (Teleostei: Osphronemidae) from Kalimantan Tengah (Borneo, Indonesia) Vertebr. Zool. 2013;63:35–40. [Google Scholar]
- Schindler I., Schmidt J. Review of the mouthbrooding Betta (Teleostei, Osphronemidae) from Thailand, with descriptions of two new species. Z. Fischkunde. 2006;8:47–69. [Google Scholar]
- Schindler I., Schmidt J. Betta kuehnei, a new species of fighting fish (Teleostei, Osphronemidae) from the Malay Peninsula. Bull. Fish Biol. 2008;10:34–96. [Google Scholar]
- Smith H.M. United States Government Printing Office; Washington, D.C.: 1945. The fresh-water fishes of Siam, or Thailand. [Google Scholar]
- Sriwattanarothai N., Steinke D., Ruenwongsa P., Hanner R., Panijpan B. Molecular and morphological evidence supports the species status of the Mahachai fighter Betta sp. Mahachai and reveals new species of Betta from Thailand. J. Fish Biol. 2010;77:414–424. doi: 10.1111/j.1095-8649.2010.02715.x. [DOI] [PubMed] [Google Scholar]
- Suvatti C. Department of Fisheries; Bangkok: 1950. Fauna of Thailand. [Google Scholar]
- Suvatti C. Royal Institute Thailand; Bangkok: 1981. Fishes of Thailand. [Google Scholar]
- Tan H.H. Redescription of Betta anabatoides Bleeker; and a new species of Betta from West Kalimantan, Borneo (Teleostei: Osphronemidae) Zootaxa. 2009;2165:59–68. [Google Scholar]
- Tan H.H. Betta pardalotos, a new species of fighting fish (Teleostei: Osphronemidae) from Sumatra, Indonesia. Raffles Bull. Zool. 2009;57:501–504. [Google Scholar]
- Tan H.H. The identity of Betta rubra (Teleostei: Osphronemidae) revisited, with description of a new species from Sumatra, Indonesia. Raffles Bull. Zool. 2013;61:323–330. [Google Scholar]
- Tan H.H., Ng P.K.L. The fighting fishes (Teleostei: Osphronemidae: genus Betta) of Singapore, Malaysia and Brunei. Raffles Bull. Zool. 2005;(Suppl. 13):43–99. [Google Scholar]
- Tanpitayacoop C., Na-Nakorn U. Proceedings of the 43rd Kasetsart University Annual Conference. 2005. Genetic variation of Betta spp. in Thailand by random amplified polymorphic DNA (RAPD) method; pp. 185–192. [Google Scholar]
- Taylor H.R., Harris W.E. An emergent science on the brink of irrelevance: a review of the past 8 years of DNA barcoding. Mol. Ecol. Resour. 2012;12:377–388. doi: 10.1111/j.1755-0998.2012.03119.x. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidthayanon C., Karnasuta J., Nabhitabhata J. Office of Environmental Policy and Planning; Bangkok: 1997. Diversity of freshwater fishes in Thailand. [Google Scholar]
- Voris H.K. Maps of Pleistocene sea levels in Southeast Asia: shorelines, river systems, time durations. J. Biogeogr. 2000;27:1153–1167. [Google Scholar]
- Ward R.D., Zemlak T.S., Innes B.H., Last P.R., Hebert P.D.N. DNA barcoding Australia's fish species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005;360:1847–1857. doi: 10.1098/rstb.2005.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward R.D., Hanner R., Hebert P.D.N. The campaign to DNA barcode all fishes, FISH-BOL. J. Fish Biol. 2009;74:329–356. doi: 10.1111/j.1095-8649.2008.02080.x. [DOI] [PubMed] [Google Scholar]
- Witte K., Schmidt J. Betta brownorum, a new species of anabantoids (Teleostei; Belontiidae) from northwestern Borneo, with a key to the genus. Ichthyol. Explor. Freshwat. 1992;2:305–330. [Google Scholar]
- Woodruff D.S. Biogeography and conservation in Southeast Asia: how 2.7 million years of repeated environmental fluctuations affect today's patterns and the future of the remaining refugial-phase biodiversity. Biodivers. Conserv. 2010;19:919–941. [Google Scholar]
- Xie W., Lewis P.O., Fan Y., Kuo L., Chen M.H. Improving marginal likelihood estimation for Bayesian phylogenetic model selection. Syst. Biol. 2011;60:150–160. doi: 10.1093/sysbio/syq085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemlak T.S., Ward R.D., Connell A.D., Holmes B.H., Hebert P.D.N. DNA barcoding reveals overlooked marine fishes. Mol. Ecol. Resour. 2009;9(Suppl. 1):237–242. doi: 10.1111/j.1755-0998.2009.02649.x. [DOI] [PubMed] [Google Scholar]